Recently, we developed novel genetic tools for precise genome editing in Ogataea (Hansenula) polymorphaby enhancing high homologous recombination (HR) rates and revealed a strong competition between HR and non-homology end joining (NHEJ). With this efficient tool, O. polymorphawas harnessed for the first production of fatty alcohols from both glucose and methanol.
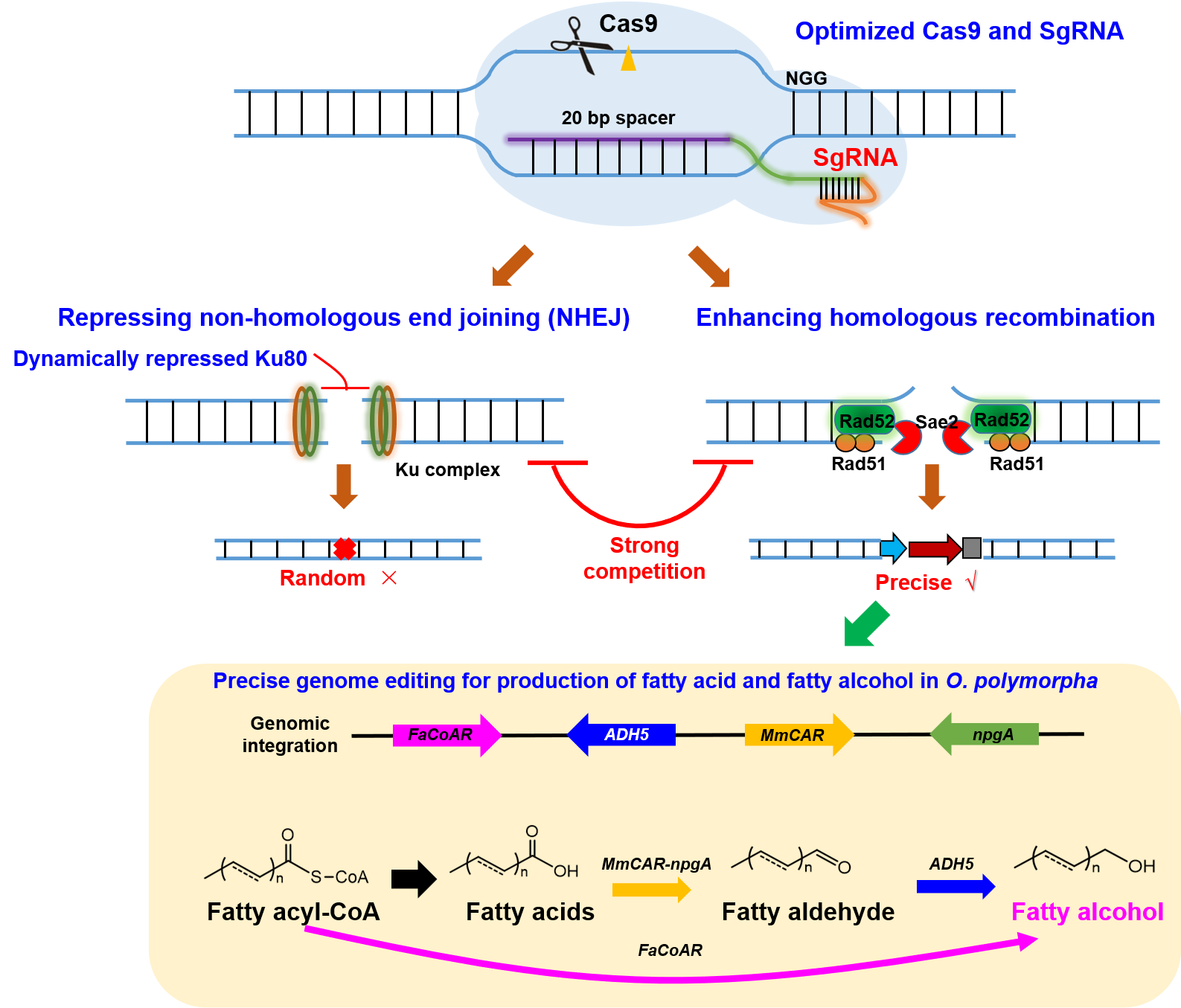
Methanol is an ideal the bio-refinery feedstock that can be served as supplements to traditional feedstocks like biomass and crops, and also contributes to the clean utilization of coal and natural gas. The thermal-tolerant O. polymorphacan efficiently assimilates methanol, and thus is considered as a promising chassis host for efficient methanol biotransformation. However, the difficulties in convenient and precise genome editing in O. polymorpha limits its metabolic engineering toward industrial application. This study constructed and optimized CRISPR/Cas9 system by fine regulation and expression of Cas9 and sgRNA, which achieved efficient genome edition with positive rates up to 90%. Then dynamically regulation of key gene KU80 in NHEJ pathway and enhancing the expression of HR-related proteins (Rad51, Rad52, Sae2), increased HR rates to 60~70%. This HR up-regulated systems has been applied for scarless gene deletion, homologous integration of large fragments and in vivo assembly of multiple fragments, which enables the first production of fatty alcohols in O. polymorpha. (Text and image by Jiaoqi Gao)
中文版

